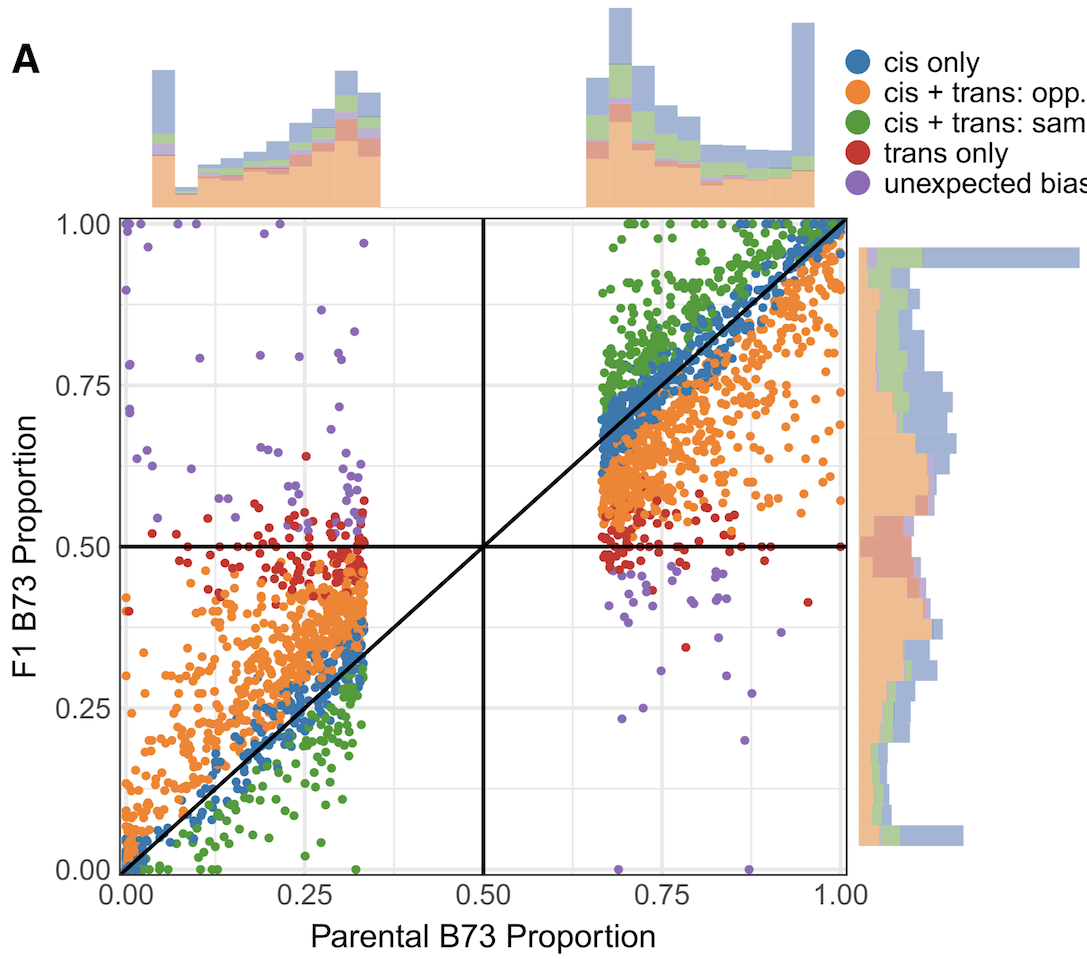
Gene expression variation is a key component underlying phenotypic variation and heterosis. Transcriptome profiling was performed on 23 different tissues or developmental stages of two maize inbreds, B73 and Mo17, as well as the F1 hybrid. This dataset provided opportunities to monitor the developmental dynamics of differential expression, additivity for gene expression, and regulatory variation. The transcriptome can be divided into ∼30,000 genes that are expressed in at least one tissue of one inbred and an additional ∼10,000 “silent” genes that are not expressed in any tissue of any genotype, 90% of which are non-syntenic relative to other grasses. Many (∼74%) of the expressed genes exhibit differential expression in at least one tissue. However, the majority of genes with differential expression do not exhibit consistent differential expression in different tissues. These genes often exhibit tissue-specific differential expression with equivalent expression in other tissues and in many cases they switch the directionality of differential expression in different tissues. This suggests widespread variation for tissue-specific regulation of gene expression between the two maize inbreds B73 and Mo17. Nearly 5,000 genes are expressed in only one parent in at least one tissue (single parent expression) and 97% of these genes are expressed at mid-parent levels or higher in the hybrid, providing extensive opportunities for hybrid complementation in heterosis. In general, additive expression patterns were much more common than non-additive patterns and this trend was emphasized for genes with strong differential expression or single parent expression. There is relatively little evidence for non-additive patterns of expression that are maintained in multiple tissues. The analysis of allele-specific expression allowed classification of cis- and trans-regulatory variation. Genes with cis-regulatory variation often exhibit additive expression and tended to have more consistent regulatory variation throughout development. In contrast, genes with trans-regulatory variation were enriched for non-additive patterns and often showed tissue-specific differential expression. Taken together, this study provides a deeper understanding of regulatory variation and the degree of additive gene expression throughout maize development. The dynamic nature of differential expression, additivity, and regulatory variation imply abundant variability for tissue-specific regulatory mechanisms, and suggest that connections between transcriptome and phenome will require expression data from multiple tissues.
Dynamic Patterns of Gene Expression Additivity and Regulatory Variation throughout Maize Development
Zhou P, Hirsch CN, Briggs SP, Springer, NM
2018.
Molecular Plant