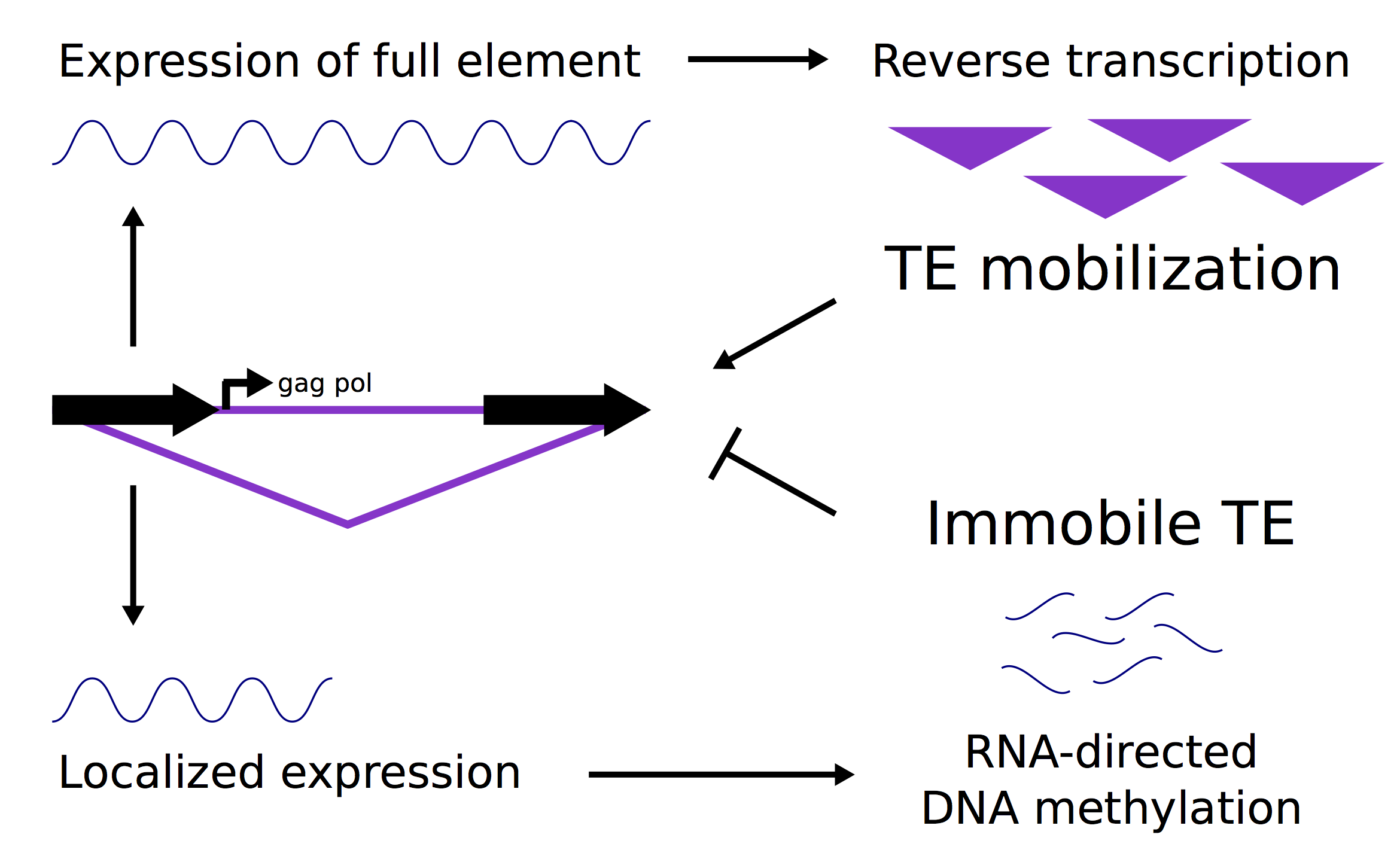
The genome of many plant species is a mosaic of genes and repetitive elements. Most of these repetitive elements are derived from different types of transposable elements (TEs). The exact types and locations of transposons present in genomes varies widely between species and even between individuals of the same species. Genomes have developed a variety of mechanisms to suppress transposon movement and to mitigate the influences of TEs on genes. Maize provides a good system for studying how TEs and genes interact since over 80% of the maize genome is derived from TEs and most genes have transposons located very nearby. We are studying how the genome uses chromatin modifications to separate genes and transposons. In addition, we aim to understand how TEs are expressed and how transposons might create new regulatory information that can influence expression of genes. This work has been supported by the National Science Foundation and the United States Department of Agriculture.